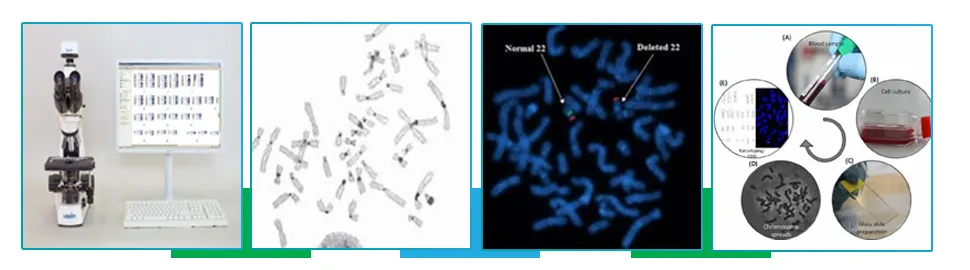
Cytogenetics Technology
Cytogenetics - the study of chromosomes which are long strands of DNA and protein that contains most of the genetic information in a cell. It involves testing of samples like tissue, blood, bone marrow, amniotic fluid etc, to look for any changes in the chromosomes, including broken, missing, rearranged or extra chromosomes. These changes in the chromosome may be a sign of a genetic disease or even certain types of cancer. Advanced equipments and methodologies such as Karyotyping, Giemsa staining are used to visualise the bands, as each chromosome has unique banding pattern. Cytogenetics takes advantage of the possibility of making the whole genome visible to human eye under the microscope. It helps in the diagnosis and classification of the disease, planning treatment regimens and monitoring status of the disease.
PCR and Real-Time PCR
Polymerase chain reaction (PCR) has completely revolutionized the detection of RNA & DNA.It is a process to amplify a single DNA molecule into thousands of copies in a matter of hours. Used for Qualitative and Quantitative analysis. The double stranded, helical DNA of interest is heated along certain enzymes to separate into two single strands.Specific sets of primers are used which bind or attach to the target gene or region. Accurate and sensitive technologies like Real-Time PCR minimizes time and helps to provide immediate information for multiple samples. Fluorescent dyes or DNA probes containing a fluorophore, such as Taqman in Real Time PCR aids to measure the amount of amplified color product in real time. Using the closed tube method for PCR’s reduces the risk of cross contamination with accurate results.

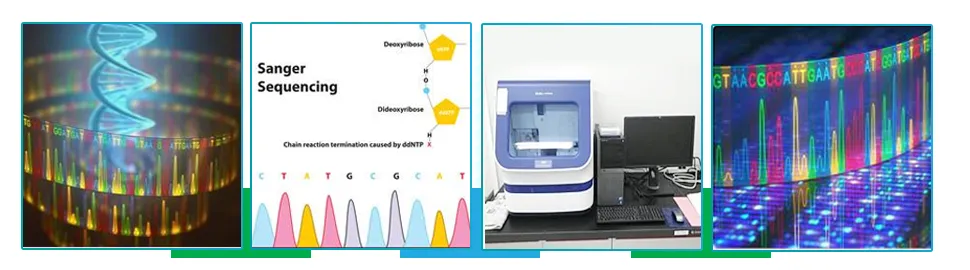
Sanger Sequencing
Sanger sequencing also known as chain termination sequencing or Dideoxy sequencing has been the gold standard for many years and, like many other assays, is based on the dideoxy-chemistry. It is a targeted sequencing technique that uses oligonucleotide primers to seek out specific DNA regions. This method has been extensively used to advance the field of functional and comparative genomics, evolutionary genetics and complex disease research. Use of this sequencing technology helps to target smaller genomic regions in larger number of samples, sequencing of variable regions, identifying single disease causing genetic variants. Its greatest advantages includes is that it is cost efficient, simultaneous interrogation of more than 100 genes at a time.
Chromosomal microarray (CMA)
Chromosomal Microarray is a microchip-based testing platform that allows high-volume, automated analysis of many pieces of DNA at once.It looks (screens) for extra (duplicated) or missing (deleted) chromosomal segments, called as copy number variants (CNVs).CMA chips use labels or probes that bind to( hybridize with) specific chromosomal regions to detect Copy number variations (CNV’s). Computer analysis is then used to compare a patient’s genetic material to that of a reference sample. A difference between a patient’s DNA and the reference sample is called a variant.CMA testing is useful for individuals who shows features not typically related to an established diagnosis, but demonstrate developmental delay/intellectual impairment, autism spectrum disorders, multiple congenital anomalies, including dysmorphic facial features.
.webp)
Fluorescence In situ hybridization (FISH)
Fluorescence in situ hybridization (abbreviated FISH) is a technique used to detect and locate a specific DNA sequence on a chromosome. In this technique, the full set of chromosomes from an individual is affixed onto a glass slide and then exposed to a probe. The fluorescently labeled probe then binds to its matching (complementary) sequence within the set of chromosomes. The chromosome and sub-chromosomal location where the fluorescent probe is bound can be visualized using a special microscope (fluorescent microscope/confocal microscope). This may be used for understanding a variety of chromosomal abnormalities that include different characteristic gene fusions in a cell, loss of a chromosomal region or a whole chromosome and other genetic mutations.
.webp)
Next Generation Sequencing (NGS)
Next Generation Sequencing is a technology used to determine the sequence of DNA or RNA to study various genetic variations.The basic steps in next-generation sequencing involves fragmenting DNA/RNA into multiple pieces, adding adapters, sequencing the libraries, and reassembling them to form a genomic sequence.This technology could aid in studying gene expression, epigenetic changes, and molecular analysis to understand a variety of diseases and aid in development of personalized medicine. The NGS technology is used to sequence human genome in order to detect genetic factors that contribute to disease, to sequence bacteria and viruses aiding to detect factors that may contribute to virulence. RNA sequencing can provide information on a sample’s entire transcriptome in one analysis, without any previous knowledge of an organism’s genetic sequence being required.
.webp)
Pathology diagnostic services for B2B & B2C customers
Pathology Diagnostic identifies the cause of disease based on clinical pathology and/or morphologic findings, It also studied on basis of history, ancillary test results and clinical signs. It is important in all areas of pathology, both in spontaneous and in experimentally-induced disease.
In experimental studies, it is important to separate out the effects of spontaneous disease and those induced by the experimental agent/test article. Diagnostic pathology is essential to investigate unexpected disease or death in laboratory animal colonies or prior to the termination of a study.

